Efficient validity checking in monadic predicate logic
Monadic predicate logic (with identity) is decidable. (See Boolos, Burgess, and Jeffrey 2007, Ch. 21. The result goes back to Löwenheim-Skolem 1915).
How can we write a program to check whether a formula is logically valid (and hence also a theorem)?
First, we have to parse the formula, meaning to convert it form a string into a format that represents its syntax in a machine-readable way. That format is an abstract syntax tree like this:
Formula: ∀x(Ax→(Ax∧Bx)) Abstract syntax tree: ∀ ├── x └── → ├── A │ └── x └── ∧ ├── A │ └── x └── B └── x
Writing the parser was a fun lesson in a fundamental aspect of computer science. But there was nothing novel about this exercise, and not much interesting to say about it.
The focus of this post, instead, is the part of the program that actually checks whether this syntax tree represents a logically valid formula.
To start with, we might try to evaluate the formula under every possible model of a given size. How big does the model need to be?
We can make use of the Löwenheim-Skolem theorem (looking first at the case without identity):
If a sentence of monadic predicate logic (without identity) is satisfiable, then it has a model of size no greater than , where is the number of predicates in the sentence. (Lemma 21.8 BBJ).
A sentence’s negation is satisfiable if and only if the sentence is not valid, so the theorem equivalently states: a sentence is valid iff it is true under every model of size no greater than .
For a sentence with predicates, every constant in the model is assigned a list of truth-values, representing for each predicate whether . We can use itertools to find every possible such list, i.e. every possible assignment to a constant.
>>> k = 2
>>> possible_predicate_combinations = [i for i in itertools.product([True,False],repeat=k)]
[(True, True), (True, False), (False, True), (False, False)]
The list of every possible assignment to a constant has a length of .
We can then ask itertools to give us, for a model of size , every possible combination of such lists of possible constant-assignments. We let be at most , because of the theorem.
>>> for m in range(1,2**k+1):
>>> possible_models = [i for i in itertools.product(possible_predicate_combinations,repeat=m)]
>>> print(len(possible_models),"possible models of size",m)
>>> for model in possible_models:
>>> print(list(model))
4 possible models of size 1
[(True, True)]
[(True, False)]
[(False, True)]
[(False, False)]
16 possible models of size 2
[(True, True), (True, True)]
[(True, True), (True, False)]
[(True, True), (False, True)]
[(True, True), (False, False)]
[(True, False), (True, True)]
[(True, False), (True, False)]
[(True, False), (False, True)]
[(True, False), (False, False)]
[(False, True), (True, True)]
[(False, True), (True, False)]
[(False, True), (False, True)]
[(False, True), (False, False)]
[(False, False), (True, True)]
[(False, False), (True, False)]
[(False, False), (False, True)]
[(False, False), (False, False)]
64 possible models of size 3
[(True, True), (True, True), (True, True)]
[(True, True), (True, True), (True, False)]
[(True, True), (True, True), (False, True)]
[(True, True), (True, True), (False, False)]
[(True, True), (True, False), (True, True)]
[(True, True), (True, False), (True, False)]
[(True, True), (True, False), (False, True)]
[(True, True), (True, False), (False, False)]
[(True, True), (False, True), (True, True)]
[(True, True), (False, True), (True, False)]
...
256 possible models of size 4
[(True, True), (True, True), (True, True), (True, True)]
[(True, True), (True, True), (True, True), (True, False)]
[(True, True), (True, True), (True, True), (False, True)]
[(True, True), (True, True), (True, True), (False, False)]
[(True, True), (True, True), (True, False), (True, True)]
[(True, True), (True, True), (True, False), (True, False)]
[(True, True), (True, True), (True, False), (False, True)]
[(True, True), (True, True), (True, False), (False, False)]
[(True, True), (True, True), (False, True), (True, True)]
[(True, True), (True, True), (False, True), (True, False)]
...
What’s unfortunate here is that for our -predicate sentence, we will need to check models. The sum is very roughly equal to its last term, . For , this is a number in the billions, for , it’s a number with 19 zeroes.
So checking every model is computationally impossible in practice. Fortunately, we can do better.
Let’s look back at the Löwenheim-Skolem theorem and try to understand why appears in it:
If a sentence of monadic predicate logic (without identity) is satisfiable, then it has a model of size no greater than , where is the number of predicates in the sentence. (Lemma 21.8 BBJ).
As we’ve seen, is the number of possible combinations of predicates that can be true of a constant in the domain. Visually, this is the number of subsets in a partition of the possibility space:
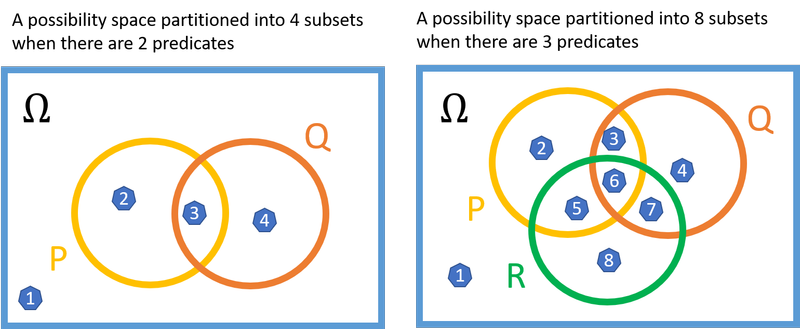
If a model had a size of, say, , one of the subsets in the partition would need to contain more than one element. But this additional element would be superfluous insofar as the truth-value of the sentence is concerned. The partition subset corresponds to a predicate-combination that would already be true with just one element in the subset, and will continue to be true if more elements are added. Take, for example, the subset labeled ‘8’ in the drawing, which corresponds to . The sentence is true whether there are one, two, or a million elements in subset 8. Similarly, does not depend on the number of elements in subset 8.
Seeing this not only illuminates the theorem, but also let us see that the vast majority of the multitudinous models we considered earlier are equivalent. All that matters for our sentence’s truth-value is whether each of the subsets is empty or non-empty. This means there are in fact only model equivalence classes to consider. We need to subtract one because the subsets cannot all be empty, since the domain needs to be non-empty.
>>> k = 2
>>> eq_classes = [i for i in itertools.product(['Empty','Non-empty'],repeat=2**k)]
>>> eq_classes.remove(('Empty',)*k**2)
>>> eq_classes
[('Empty', 'Empty', 'Empty', 'Non-empty'),
('Empty', 'Empty', 'Non-empty', 'Empty'),
('Empty', 'Empty', 'Non-empty', 'Non-empty'),
('Empty', 'Non-empty', 'Empty', 'Empty'),
('Empty', 'Non-empty', 'Empty', 'Non-empty'),
('Empty', 'Non-empty', 'Non-empty', 'Empty'),
('Empty', 'Non-empty', 'Non-empty', 'Non-empty'),
('Non-empty', 'Empty', 'Empty', 'Empty'),
('Non-empty', 'Empty', 'Empty', 'Non-empty'),
('Non-empty', 'Empty', 'Non-empty', 'Empty'),
('Non-empty', 'Empty', 'Non-empty', 'Non-empty'),
('Non-empty', 'Non-empty', 'Empty', 'Empty'),
('Non-empty', 'Non-empty', 'Empty', 'Non-empty'),
('Non-empty', 'Non-empty', 'Non-empty', 'Empty'),
('Non-empty', 'Non-empty', 'Non-empty', 'Non-empty')]
We are now ready to consider the extension to monadic predicate logic with identity. With identity, it’s possible to check whether any two members of a model are distinct or identical. This means we can distinguish the case where a partition subset contains one element from the case where it contains several. But we can still only distinguish up to a certain number of elements in a subset. That number is bounded above by the number of variables in the sentence1 (e.g. if you only have two variables and , it’s not possible to construct a sentence that asserts there are three different things in some subset). Indeed we have:
If a sentence of monadic predicate logic with identity is satisfiable, then it has a model of size no greater than , where is the number of monadic predicates and the number of variables in the sentence. (Lemma 21.9 BBJ)
By analogous reasoning to the case without identity, we need only consider model equivalence classes. All that matters for our sentence’s truth-value is whether each of the subsets has or elements in it.
>>> k = 2
>>> r = 2
>>> eq_classes = [i for i in itertools.product(range(r+1),repeat=2**k)]
>>> eq_classes.remove((0,)*k**2)
>>> eq_classes
[(0, 0, 0, 1),
(0, 0, 0, 2),
(0, 0, 1, 0),
(0, 0, 1, 1),
(0, 0, 1, 2),
(0, 0, 2, 0),
(0, 0, 2, 1),
(0, 0, 2, 2),
(0, 1, 0, 0),
(0, 1, 0, 1),
(0, 1, 0, 2),
(0, 1, 1, 0),
(0, 1, 1, 1),
...
-
I believe it should be possible to find a tighter bound based on the number of times the equals sign actually appears in the sentence. For example, if equality is only used once, e.g. in where does not contain equality, it seems clear that the number of variables in should have no bearing on the model size that is needed. My hunch is that more generally you need uses of ‘’ to assert that objects are distinct, so, for example if ‘’ appears 5 times you can distinguish 3 objects in a subset, or with 12 ‘’s you can distinguish 5 objects. It’s only an intuition and I haven’t checked it carefully. ↩